Microbes Isolated from Landfill Soil Utilize Polyethylene Terephthalate (PET) as Their Sole Source of Carbon: An Unexplored Possibility of Bioremediation in Bangladesh 10.32526/ennrj/22/20230124
Main Article Content
Abstract
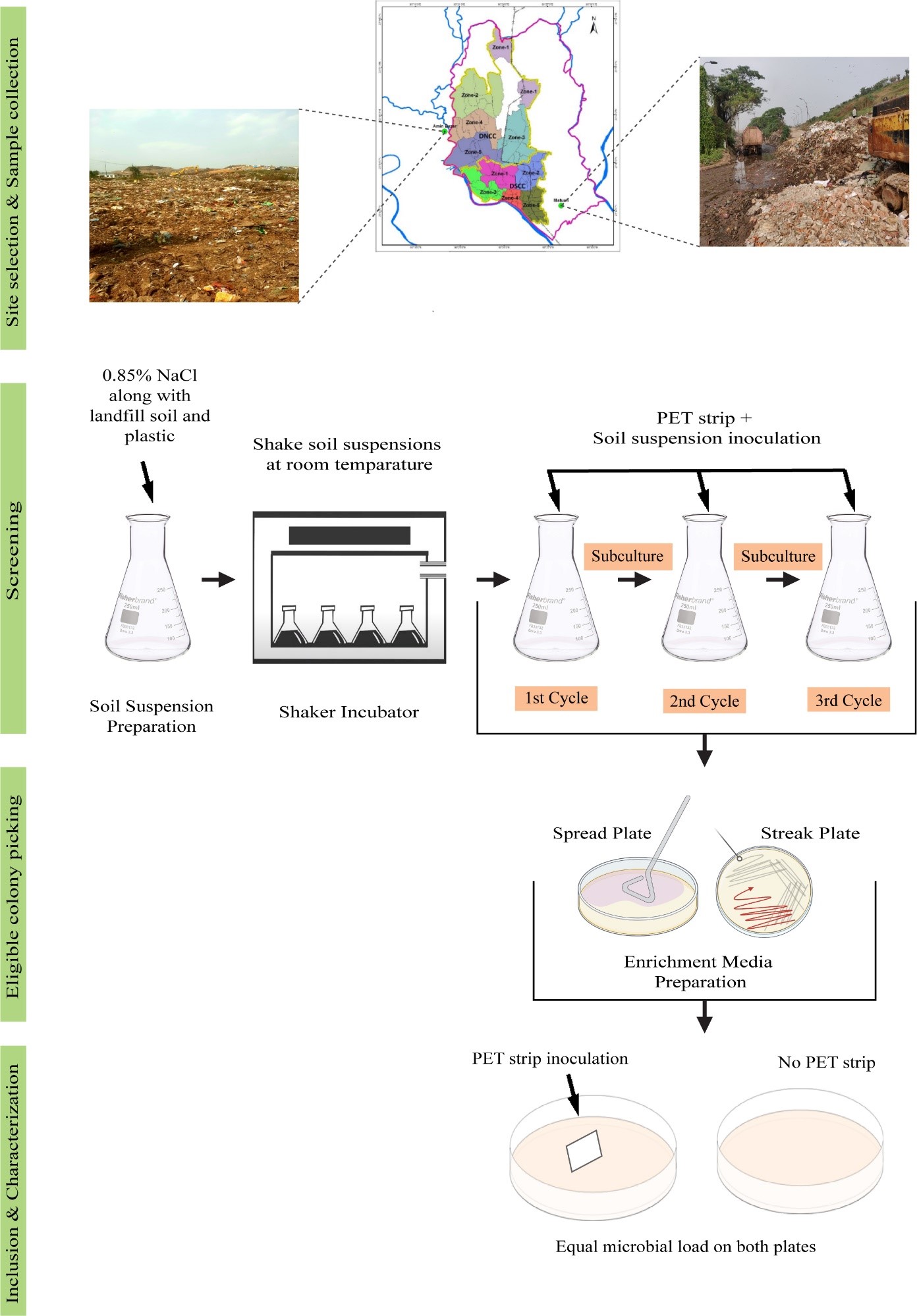
Plastic products are so extensively used that they continue to strain the already overburdened waste management system and, inevitably, the global climate. Biodegradation is a sustainable remedy. Here, we report a few microorganisms isolated from landfill soil near Dhaka that thrive especially on polyethylene terephthalate (PET) polymers. Soil samples were subjected to three enrichment cycles that contained no carbon except PET. Pure isolates were recovered and incubated on minimal agar containing PET as the sole carbon. A morphological examination was carried out. Potential PET-degrading enzyme sequences from the isolates and other microalgae were analyzed for homology using BLASTP and TBLASTN, and multiple sequence alignment (MSA) was performed to assess conserved domains. Six isolates were obtained. Two isolates grew around the PET film but did not grow sufficiently in other areas of the minimal agar. Two other isolates with greenish pigmentation flourished around the PET film as well as on other areas of the agar. One of the green cells resembled Aphanocapsa, with irregular shapes and occasionally brown dense bodies, while the others looked round like Microcystis. Homology analysis revealed the hypothetical PETases in green cells contained the highly conserved catalytic triad (Ser-His-Asp) at the active site, as always found in alpha-beta hydrolase fold containing enzymes. Microbes isolated from two landfill sites in the vicinity of Dhaka have been adapted to utilize PET as a carbon source. In the future, sequencing and further characterization would be necessary to validate the findings. Microalgal systems demand increased focus, given their potential to offer valuable resources for bioremediation.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Published articles are under the copyright of the Environment and Natural Resources Journal effective when the article is accepted for publication thus granting Environment and Natural Resources Journal all rights for the work so that both parties may be protected from the consequences of unauthorized use. Partially or totally publication of an article elsewhere is possible only after the consent from the editors.
References
Akram M, Khan MA, Ahmed N, Bhatti R, Pervaiz R, Malik K, et al. Cloning and expression of an anti-cancerous cytokine: Human IL-29 gene in Chlamydomonas reinhardtii. AMB Express 2023;13:Aticle No. 23.
Akter S, Shammi M, Jolly YN, Sakib AA, Rahman MM, Tareq SM. Characterization and photodegradation pathway of the leachate of Matuail sanitary landfill site, Dhaka South City Corporation, Bangladesh. Heliyon 2021;7(9):e07924.
Almeida EL, Carrillo Rincόn AF, Jackson SA, Dobson ADW. In silico screening and heterologous expression of a polyethylene terephthalate hydrolase (PETase)-like enzyme (SM14est) with polycaprolactone (PCL)-degrading activity, from the marine sponge-derived strain Streptomyces sp. SM14. Frontiers in Microbiology 2019;10:Article No. 2187.
Austin HP, Allen MD, Donohoe BS, Rorrer NA, Kearns FL, Silveira RL, et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proceedings of the National Academy of Sciences of the United States of America 2018;115(19):4350-7.
Badr AA, Fouad WM. Identification of culturable microalgae diversity in the River Nile in Egypt using enrichment media. African Journal of Biological Sciences 2021;3(2):Article No. 50.
Barnes DKA, Galgani F, Thompson RC, Barlaz M. Accumulation and fragmentation of plastic debris in global environments. Philosophical Transactions of the Royal Society B 2009; 364:1985-98.
Barone GD, Ferizovi´ D, Biundo A, Lindblad P. Hints at the applicability of microalgae and Cyanobacteria for the biodegradation of plastics. Sustainability 2020;12(24):Article No. 10449.
Barth M, Honak A, Oeser T, Wei R, Belisário-Ferrari MR, Then J, et al. A dual enzyme system composed of a polyester hydrolase and a carboxylesterase enhances the biocatalytic degradation of polyethylene terephthalate films. Biotechnology Journal 2016;11(8):1082-7.
Bellinger EG, Sigee DC. Freshwater Algae: Identification, Enumeration and Use as Bioindicators. 2nd ed. John Wiley and Sons; 2015.
Biki SP, Mahmud S, Akhter S, Rahman Md J, Rix JJ, Bachchu Md AA, et al. Polyethylene degradation by Ralstonia sp. strain SKM2 and Bacillus sp. strain SM1 isolated from land fill soil site. Environmental Technology and Innovation 2021; 22:Article No. 101495.
Biovia DS. Discovery Studio Visualizer [Internet]. 2021 [cited 2022 Dec 3]. Available from: https://discover.3ds.com/ discovery-studio-visualizer-download.
Brott S, Pfaff L, Schuricht J, Schwarz JN, Böttcher D, Badenhorst CPS, et al. Engineering and evaluation of thermostable IsPETase variants for PET degradation. Engineering in Life Sciences 2022;22(3-4):192-203.
Carniel A, Valoni É, Junior JN, Gomes ADC, Castro AMD. Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergistically for PET hydrolysis to terephthalic acid. Process Biochemistry 2017;59:84-90.
Carr CM, Clarke DJ, Dobson ADW. Microbial polyethylene terephthalate hydrolases: Current and future perspectives. Frontiers in Microbiology 2020;11:Article No. 571265.
Chandan MSK. A tale of a landfill and its ravages [Internet]. 2021 [cited 2022 Feb 12]. Available from: https://www.thedailystar. net/news/bangladesh/news/tale-landfill-and-its-ravages-2144066.
Chia WY, Tang DYY, Khoo KS, Lup ANK, Chew KW. Nature’s fight against plastic pollution: Algae for plastic biodegradation and bioplastics production. Environmental Science and Ecotechnology 2020;4:Article No. 100065.
Consortium TU. UniProt: The universal protein knowledgebase in 2023. Nucleic Acids Research 2023;51(D1):523-31.
Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods in Molecular Biology 2015; 1263:243-50.
Danso D, Schmeisser C, Chow J, Zimmermann W, Wei R, Leggewie C, et al. New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Applied and Environmental Microbiology 2018;84(8): e02773-17.
Demirkan E, Güler BE, Sevgi T. Analysis by scanning electron microscopy of polyethylene terephthalate and nylon biodegradation abilities of Bacillus sp. strains isolated from soil. Journal of Biological and Environmental Sciences 2020;14(42):107-14.
Dong Q, Xing X. Chroococcidiorella tianjinensis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta), a green alga arises from the Cyanobacterium TDX16. American Journal of Plant Sciences 2020;11(11):1814-26.
Falah W, Chen FJ, Zeb BS, Hayat MT, Mahmood Q, Ebadi A, et al. Polyethylene terephthalate degradation by microalga Chlorella vulgaris along with pretreatment. Materiale Plastice 2020;57(3):260-70.
Felisberto S, Souza D. Characteristics and diversity of Cyanobacteria in periphyton from lentic tropical ecosystem, Brazil. Advances in Microbiology 2014;4(15):1076-87.
Gama WJ, Laughinghouse HD, St Anna CL. How diverse are coccoid cyanobacteria? A case study of terrestrial habitats from the Atlantic Rainforest (Sao Paulo, Brazil). Phytotaxa 2014;178(2):61-97.
Gamerith C, Vastano M, Ghorbanpour SM, Zitzenbacher S, Ribitsch D, Zumstein MT, et al. Enzymatic degradation of aromatic and aliphatic polyesters by P. pastoris expressed cutinase 1 from Thermobifida cellulosilytica. Frontiers in Microbiology 2017;8:Article No. 938.
Guillard RRL, Keller MD, O’Kelly CJ, Floyd GL. Pycnococcus provasolii gen. et sp. nov., a coccoid prasinoxanthin-containing phytoplankter from the Western North Atlantic and Gulf of Mexico. Journal of Phycology 1991;27(1):39-47.
Han X, Liu W, Huang JW, Ma J, Zheng Y, Ko TP, et al. Structural insight into catalytic mechanism of PET hydrolase. Nature Communications 2017;8:Article No. 2106.
Hempel F, Maier UG. Microalgae as solar-powered protein factories. Advances in Experimental Medicine and Biology 2016;896:241-62.
Hossain KS, Das S, Kundu S, Afrin S, Nurunnabi TR, Rahman SMM. Isolation and characterization of polythene degrading bacteria from garbage soil. International Journal of Agriculture, Environment and Bioresearch 2019;4(5):254-63.
Jangra MR, Nehra KS, Garima, Kajal, Monika, Sonia, et al. Assessment of plastic degrading ability of microbes isolated from local plastic dumping sites. Bioscience Biotechnology Research Communications 2020;13(4):1668-72.
Joo S, Cho IJ, Seo H, Son HF, Sagong HY, Shin TJ, et al. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nature Communications 2018; 9:Article No. 382.
Kawai F, Oda M, Tamashiro T, Waku T, Tanaka N, Yamamoto M, et al. A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Applied Microbiology and Biotechnology 2014;98(24):10053-64.
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 2015;10:845-58.
Kennedy J, Baker P, Piper C, Cotter PD, Walsh M, Mooij MJ, et al. Isolation and analysis of bacteria with antimicrobial activities from the marine sponge Haliclona simulans collected from Irish Waters. Marine Biotechnology 2009;11:384-96.
Kim JW, Park SB, Tran QG, Cho DH, Choi DY, Lee YJ, et al. Functional expression of polyethylene terephthalate-degrading enzyme (PETase) in green microalgae. Microbial Cell Factories 2020;19:Article No. 97.
Kumar RV, Kanna GR, Elumalai S. Biodegradation of polyethylene by green photosynthetic microalgae. Journal of Bioremediation and Biodegradation 2017;8(1):Article No. 381.
Malafatti-Picca L, Chaves MRDB, Castro AMD, Valoni É, Oliveira VMD, Marsaioli AJ, et al. Hydrocarbon-associated substrates reveal promising fungi for poly (ethylene terephthalate) (PET) depolymerization. Brazilian Journal of Microbiology 2019;50(3):633-48.
Maurya A, Bhattacharya A, Khare SK. Enzymatic Remediation of polyethylene terephthalate (PET)-based polymers for effective management of plastic wastes: An overview. Frontiers in Bioengineering and Biotechnology 2020;8:Article No. 602325.
Mohanan N, Montazer Z, Sharma PK, Levin DB. Microbial and enzymatic degradation of synthetic plastics. Frontiers in Microbiology 2020;11:Article No. 580709.
Moog D, Schmitt J, Senger J, Zarzycki J, Rexer KH, Linne U, et al. Using a marine microalga as a chassis for polyethylene terephthalate (PET) degradation. Microbial Cell Factories 2019;18:Article No. 171.
Najeeb MI, Ahmad MD, Anjum AA, Maqbool A, Ali MA, Nawaz M, et al. Distribution, screening and biochemical characterization of indigenous microalgae for bio-mass and bio-energy production potential from three districts of Pakistan. Brazilian Journal of Biology 2022;84:e261698.
Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, et al. The α/β hydrolase fold. Protein Engineering 1992; 5(3):197-211.
Palm GJ, Reisky L, Böttcher D, Müller H, Michels EAP, Walczak MC, et al. Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nature Communications 2019;10:Article No. 1717.
Park H, Park J, Lin SH, Boorady LM. Assessment of Firefighters’ needs for personal protective equipment. Fashion and Textiles 2014;1:Article No. 8.
Pei J, Kim BH, Grishin NV. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Research 2008;36(7):2295-300.
Pereira VL, Portillo ER. Isolation, culture and morphological characterization of Microcystis sp. toxic strain from the tacuary reservoir. International Journal of Advanced Research 2018; 6:387-93.
Pham TL, Tran THY, Shimizu K, Li Q, Utsumi M. Toxic cyanobacteria and microcystin dynamics in a tropical reservoir: Assessing the influence of environmental variables. Environmental Science and Pollution Research 2021;28(45):63544-57.
Qi X, Yan W, Cao Z, Ding M, Yuan Y. Current advances in the biodegradation and bioconversion of polyethylene terephthalate. Microorganisms 2021;10(1):Article No. 39.
Read BA, Kegel J, Klute MJ, Kuo A, Lefebvre SC, Maumus F, et al. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 2013;499:209-13.
Rolfe MD, Rice CJ, Lucchini S, Pin C, Thompson A, Cameron ADS, et al. Lag phase is a distinct growth phase that prepares bacteria for exponential growth and involves transient metal accumulation. Journal of Bacteriology 2012;194(3):686-701.
Rüthi J, Cerri M, Brunner I, Stierli B, Sander M, Frey B. Discovery of plastic-degrading microbial strains isolated from the alpine and Arctic terrestrial plastisphere. Frontiers in Microbiology 2023;14:Article No. 1178474.
Samak NA, Jia Y, Sharshar MM, Mu T, Yang M, Peh S, et al. Recent advances in biocatalysts engineering for polyethylene terephthalate plastic waste green recycling. Environment International 2020;145:Article No. 106144.
Silva C, Da S, Silva N, Matamá T, Araújo R, Martins M, et al. Engineered Thermobifida fusca cutinase with increased activity on polyester substrates. Biotechnology Journal 2011;6(10):1230-9.
Skariyachan S, Megha M, Kini MN, Mukund KM, Rizvi A, Vasist K. Selection and screening of microbial consortia for efficient and ecofriendly degradation of plastic garbage collected from urban and rural areas of Bangalore, India. Environmental Monitoring and Assessment 2015;187(1):Article No. 4174.
Sugoro I, Pikoli MR, Rahayu DS, Puspito MJ, Shalsabilla SE, Ramadhan F, et al. Microalgae diversity in interim wet storage of spent nuclear fuel in Serpong, Indonesia. International Journal of Environmental Research and Public Health 2022;19(22):Article No. 15377.
Taniguchi I, Yoshida S, Hiraga K, Miyamoto K, Kimura Y, Oda K. Biodegradation of PET: Current status and application aspects. ACS Catalysis 2019;9(5):4089-105.
Tortora GJ, Funke BR, Case CL. Microbiology: An Introduction. 9th ed. San Francisco: Pearson Benjamin Cummings; 2007.
Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry 2010;31(2):455-61.
Turmel M, Gagnon MC, O’Kelly CJ, Otis C, Lemieux C. The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of Prasinophytes and the origin of the secondary chloroplasts of euglenids. Molecular Biology and Evolution 2009;26(3):631-48.
Urbanek AK, Kosiorowska KE, Mirończuk AM. Current knowledge on polyethylene terephthalate degradation by genetically modified microorganisms. Frontiers in Bioengineering and Biotechnology 2021;9:Article No. 771133.
Urbanek AK, Rymowicz W, Strzelecki MC, Kociuba W, Franczak L, Mirończuk AM. Isolation and characterization of Arctic microorganisms decomposing bioplastics. AMB Express 2017;7:Article No. 148
Urme SA, Radia MA, Alam R, Chowdhury MU, Hasan S, Ahmed S, et al. Dhaka landfill waste practices: Addressing urban pollution and health hazards. Buildings and Cities 2021; 2(1):700-16.
Waring RH, Harris RM, Mitchell SC. Plastic contamination of the food chain: A threat to human health? Maturitas 2018;115:64-8.
Wei R, Oeser T, Schmidt J, Meier R, Barth M, Then J, et al. Engineered bacterial polyester hydrolases efficiently degrade polyethylene terephthalate due to relieved product inhibition. Biotechnology and Bioengineering 2016;113(8):1658-65.
Yang S, Xu H, Yan Q, Liu Y, Zhou P, Jiang Z. A low molecular mass cutinase of Thielavia terrestris efficiently hydrolyzes poly(esters). Journal of Industrial Microbiology and Biotechnology 2013;40(2):217-26.
Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, et al. A bacterium that degrades and assimilates poly (ethylene terephthalate). Science 2016;351(6278):1196-9.
Zhang H, Perez-Garcia P, Dierkes RF, Applegate V, Schumacher J, Chibani CM, et al. The bacteroidetes Aequorivita sp. and Kaistella jeonii produce promiscuous esterases with PET-hydrolyzing activity. Frontiers in Microbiology 2022;12: Article No. 803896.