Streptomyces sp. Strain SRH22: A Potential Bioremediation Agent for Glyphosate-Contaminated Agricultural Soils 10.32526/ennrj/21/20230181
Main Article Content
Abstract
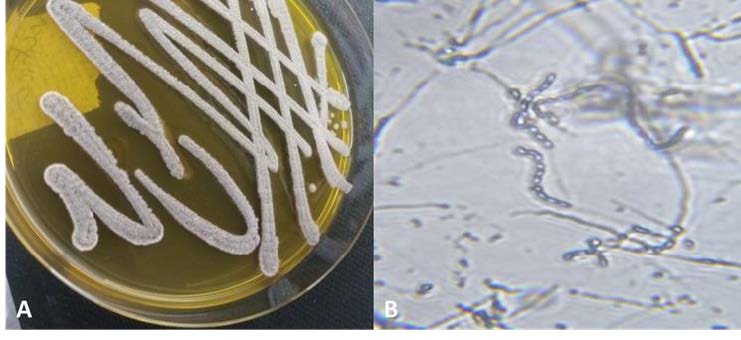
Glyphosate, also known as N-phosphonomethylglycine, is the herbicide that is widely used across the globe. As there are concerns over its potential toxicity to non-target soil species, there is a growing interest in identifying glyphosate-degrading microorganisms in soil. Biodegradation, by actinobacteria, is a very promising approach to eliminate this pesticide from contaminated environments. The present work isolated and identified actinobacteria capable of degrading glyphosate from Saharan agriculture, as well as determined how the application of this herbicide affects the abundance of actinobacteria present in soil. It was observed that the use of glyphosate led to an increased abundance of actinobacteria in the soil compared to the untreated soil. Among this population, an actinobacterial strain was isolated from glyphosate contaminated soil by the enrichment method, and was identified to possess the greatest capability to degrade glyphosate at 50 mg/L. The identification of this strain was achieved through a combination of cultural, morphological, biochemical, and molecular techniques. This included the use of 16S rDNA sequencing, leading to its successful classification as Streptomyces sp. strain SRH22. This strain was assigned the accession number OQ302556 by the National Center for Biotechnology Information (NCBI). A rapid, sensitive, and straightforward spectrophotometric technique was employed for the quantification of glyphosate. Results showed that the optimal biodegradation (90.2%) was obtained under a temperature of 30 degrees, a PH of 7.2, and an inoculum volume of 4% timed over six days. This work shows that the Streptomyces SRH22 presents good potentiality to be used as a bioremediation agent for agricultural soils in the Algerian Sahara.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Published articles are under the copyright of the Environment and Natural Resources Journal effective when the article is accepted for publication thus granting Environment and Natural Resources Journal all rights for the work so that both parties may be protected from the consequences of unauthorized use. Partially or totally publication of an article elsewhere is possible only after the consent from the editors.
References
Abosereha NA, Salima RG, El-Sayeda AF, Hammadb MA, Elsayeda GM. In-vitro biodegradation of glyphosate using genetically improved bacterial isolates from highly polluted wastewater. Egyptian Journal of Chemistry 2022; 65(132):669-81.
Abraham J, Gajendiran A. Biodegradation of fipronil and its metabolite fipronilsulfone by Streptomyces rochei strain AJAG7 and its use in bioremediation of contaminated soil. Pesticide Biochemistry and Physiology 2019;155(1):90-100.
Adelowo FE, Olu-Arotiowa OA, Amuda OS. Biodegradation of glyphosate by fungi species. Journal of Bioscience and Bioengineering 2014;2(1):104-18.
Alvarez A, Saez JS, Costa JSD, Colin VL, Fuentes MS, Cuozzo SA, et al. Actinobacteria: Current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 2016;166:41-62.
Araujo ASF, Monteiro RTR, Abarkeli RB. Effect of glyphosate on the microbial activity of two Brazilian soils. Chemosphere 2003;52(5):799-804.
Arias-Estévez M, Lopez-Periago E, Martinez-Carballo E, Simal-Gandara J, Mejuto JC, Garcia-Rio L. The mobility and degradation of pesticides in soils and the pollution of groundwater resources. Agriculture, Ecosystems and Environment 2008;123(4):247-60.
Bai SH, Ogbourne SM. Glyphosate: Environmental contamination, toxicity and potential risks to human health via food contamination. Environmental Science and Pollution Research 2016;23(19):18988-9001.
Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Klenk HP, et al. Taxonomy, physiology, and natural products of actinobacteria. Microbiology and Molecular Biology Reviews 2016;80(1):1-43.
Belhadi A, Mehenni M, Reguieg L, Yakhlef H. Phytosanitary practices of greenhouses farmers in three region of Ziban east and their potential impact on human health and environment. Revue Agricole 2016;1:9-16.
Abdelhak IM. A Geochemical Approach to Predicting the Salinity of Bare Soils Under Arid Conditions [dissertation]. Ouargla, University of Kasdi Merbah; 2020 (in French).
Benslama O, Boulahrouf A. Isolation and characterization of glyphosate-degrading bacteria from different soils of Algeria. African Journal of Microbiology Research 2013;7(49);5587-95.
Bhaskara BL, Nagaraja P. Direct sensitive spectrophotometric determination of glyphosate by using ninhydrin as a chromogenic reagent in formulations and environmental water samples. Helvetica Chimica Acta 2006;89(11):2686-93.
Bhatt P, Bhandari G, Bhatt K, Maithani D, Mishra S, Gangola S, et al. Plasmid-mediated catabolism for the removal of xenobiotics from the environment. Journal of Hazardous Materials 2021a;420(10):Article No. 126618.
Boudemagh A, Kitouni A, Boughachiche F, Hamdiken H, Oulmi L, Reghioua S, et al. Isolation and molecular identification of actinomycet microflora, of some Saharian soils of south east Algeria (Biskra, EL-Oued and Ouragla) study of antifungal activity of isolated strains. Journal of Medical Mycology 2005;15(1):39-44.
Boufercha O, Moreira IS, Castro PML, Boudemagh A. Actinobacteria isolated from wastewater treatment plants located in the east‐north of Algeria able to degrade pesticides. World Journal of Microbiology and Biotechnology 2022;38(6):Article No. 105.
Briceño G, Fuentes MS, Palma G, Jorquera MA, Amoroso MJ, Diez MC. Chlorpyrifos biodegradation and 3,5,6-trichloro-2-pyridinol production by actinobacteria isolated from soil. International Biodeterioration and Biodegradation 2012;73:1-7.
Briceño G, Fuentes MS, Saez JM, Diez M, Benimeli CS. Streptomyces genus as biotechnological tool for pesticide degradation in polluted systems. Critical Reviews in Environmental Science and Technology 2018;48:773-805.
Briceño G, Vergara K, Schalchli H, Palma G, Tortella G, Fuentes MS, et al. Organophosphorus pesticide mixture removal from environmental matrices by a soil Streptomyces mixed culture. Environmental Science and Pollution Research 2017; 25(22):21296-307.
Cardozo M, de Almeida JSFD, de Cavalcante SFA, Salgado JRS, Gonçalves AS, França TCC, et al. Biodegradation of organophosphorus compounds predicted by enzymatic process using molecular modelling and observed in soil samples through analytical techniques and microbiological analysis: A comparison. Molecules 2019;25(58):2-21.
Carlisle SM, Trevors TJ. Effect of the herbicide glyphosate on respiration and hydrogen consumption in soil. Water, Air, and Soil Pollution 1986;27:391-401.
De Castilhos Ghisi N, Zuanazzi NR, Fabrin TMC, Oliveira EC. Glyphosate and its toxicology: A scientometric review. Science of the Total Environment 2020;733:Article No. 139359.
Elarabi NI, Abdelhadi AA, Ahmed RH, Saleh I, Arif IA, Osman G, et al. Bacillus aryabhattai FACU: A promising bacterial strain capable of manipulate the glyphosate herbicide residues. Saudi Journal of Biological. Sciences 2020;27(9):2207-14.
Ermakova IT, Shushkov TV, Sviridov A, Zelenkova NF, Vinokurova NG, Baskunov BP, et al. Organophosphonates utilization by soil strains of Ochrobactrum anthropic and Achromobacter sp. Archives of Microbiology 2017;199(5): 665-75.
Firdous S, Iqbal S, Anwa S. Optimization and modeling of glyphosate biodegradation by a novel Comamonas odontotermitis P2 through response surface methodology. Pedosphere 2020;30(5):618-27.
Fuentes MS, Raimondo EE, Amoroso MJ, Benimeli CS. Removal of a mixture of pesticides by a Streptomyces consortium: Influence of different soil systems. Chemosphere 2017;173:359-67.
Gongora-Echeverría VR, García-Escalante R, Rojas-Herrera R, Giacoman-Vallejos G, Ponce-Caballero C. Pesticide bioremediation in liquid media using a microbial consortium and bacteria-pure strains isolated from a biomixture used in agricultural areas. Ecotoxicology and Environmental Safety 2020;200(5):Article No.110734.
Hadi F, Mousavi A, Noghabi KA, Tabar HG, Salmanian AH. New bacterial strain of the genus Ochrobactrum with glyphosate-degrading activity. Journal of Environmental Health 2013; 48(3):208-13.
Hernandez Guijarro K, Aparicio V, De Geronimo E, Castellote M, Figuerola EL, Costa JL, et al. Soil microbial communities and glyphosate decay in soils with different herbicide application history. Science of the Total Environment 2018;634(9):974-82.
Jiang B, Zhang N, Xing Y, Lian L, Chen Y, Zhang D, et al. Microbial degradation of organophosphorus pesticides: Novel degraders, kinetics, functional genes, and genotoxicity assessment. Environmental Science and Pollution Research 2019;26(21):21668-81.
Korayem AS, Abdelhafez AA, Zaki E, Saleh MM. Optimization of biosurfactant production by Streptomyces isolated from Egyptian arid soil using Plackett-Burman design. Annals of Agricultural Science 2015;60(2):209-17.
Kryuchkova YV, Burygin G, Gogoleva NE, Gogolev YV, Chernyshova MP, Makarov OE, et al. Isolation and caracterisation of a glyphosate-degrading rhizosphere strain, Eterobacter cloacae K7. Microbiological Research 2013; 169(1):99-105.
Kuklinsky-Sobral J, Araujo WL, Mendes R, Pizzirani-Kleiner AA, Azevedo JL. Isolation and characterization of endophytic bacteria from soybean (Glycine max) grown in soil treated with glyphosate herbicide. Plant Soil 2005;273(1-2):91-9.
Lee LH, Zainal N, Azman AS, Mutalib NSA, Hong K, Chan KG. Mumia flava gen. nov., sp. nov., an actinobacterium of the family Nocardioidaceae. International Journal of Systematic and Evolutionary Microbiology 2014;64(Pt-5):1461-7.
Li Q, Chen X, Jiang Y, Jiang C. Cultural, physiological, and biochemical identifcation of actinobacteria. In: Dhanasekaran D, Jiang Y, editors. Actinobacteria: Basics and Bio-technological Applications. InTech; 2016. p. 88-111.
Lipok J, Wieczorek D, Jewginski M, Kafarski P. Prospects of in vivo 31P NMR method in glyphosate degradation studies in whole cell system. Enzyme and Microbial Technology 2009; 44(1):11-6.
Maeda H, Dudareva N. The shikimate pathwayand aromatic amino acid biosynthesis in plants. Annual Review of Plant Biology 2012;63(1):73-105.
Malla MA, Dubey A, Kumar A, Yadav S, Kumari S. Modeling and optimization of chlorpyrifos and glyphosate biodegradation using RSM and ANN: Elucidating their degradation pathways by GC-MS based metabolomics. Ecotoxicology and Environmental Safety 2023;252(4)Article No. 114628.
Manogaran M, Shukor MY, Yasid NA, Johari WLW, Ahmad SA. Isolation and characterisation of glyphosate-degrading bacteria isolated from local soils in Malaysia. Rendiconti Lincei 2017;28(2):471-9
Minotto E, Milagre LP, Oliveira MT. Enzyme characterization of endophytic actinobacteria isolated from tomato plants. Journal of Advanced Scientific Research 2014;5(2):16-23.
Mishra S, Pang S, Zhang W, Lin Z, Bhatt P, Chen S. Insights into the microbial degradation and biochemical mechanism of carbamates. Chemosphere 2021;279(2):Article No. 130500.
Moreira IS, Lebel A, Peng X, Paula MLC, Goncalves D. Sediments in the mangrove areas contribute to the removal of endocrine disrupting chemicals in coastal sediments of Macau SAR, China, and harbour microbial communities capable of degrading E2, EE2, BPA and BPS. Biodegradation 2021;32(5):511-29.
Nnamonu LA, Nkpa NN. Use of bufers in spectrophotometric determination of N-phosphonomethylglycine by the ninhydrin colour reaction. IOSR Journal of Environmental Science Toxicology and Food Technology 2012;1(1):6-10.
Nourouzi MM, Chuah TG, Choong TSY, Rabiei F. Modeling biodegradation and kinetics of glyphosate by artificial neural network. Journal of Environmental Science and Health, Part B 2012;47(5):455-65.
Obojska A, Lejczak B, Kubrak M. Degradation of phosphonates by Streptomycete isolates. Applied Microbiology and Biotechnol 1999;51(6):872-6.
Prankle P, Meggitt W, Penner D. Rapid inactivation of glyphosate in soil. Weed Science 1975;23(3):224-8.
Reghioua S, Boughachiche F, Zerizer H, Oulmi L, Kitouni M, Boudemagh A. Antibacterial activity of rares actinomycetes isolated from arid soil samples of the southeast of Algeria. Antibiotiques 2006;8(3):147-52.
Rossi F, Carles L, Donnadieu F, Batisson I, Artigas J. Glyphosate-degrading behavior of five bacterial strains isolated from stream biofilms. Journal of Hazardous Materials 2021; 420:Article No. 126651.
Sabzevari S, Hofman J. A worldwide review of currently used pesticides monitoring in agricultural soils. Science of the Total Environment 2022;812:Article No. 152344.
Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. International Journal of Systematic Bacteriology 1966;16(3):313-40.
Singh BK, Walker A. Microbial degradation of organophosphorus compounds. FEMS Microbiology Reviews 2006;30(3):428-71.
Singh S, Kumar V, Sidhu GK, Singh J. Kinetic study of the biodegradation of glyphosate by indigenous soil bacterial isolates in presence of humic acid, Fe (III) and Cu (II) ions. Journal of Environmental Chemical Engineering 2019; 7(3):Article No.103098.
Souagui Y, Grosdemange-Billiard C, Tritsch D, Kecha M. Antifungal molecules produced by a new salt-tolerant and alkaliphilic Streptomyces sp. BS30 isolated from an arid soil. Proceedings of the National Academy of Sciences, India Section B: Biological Sciences 2015;87(2):527-35.
Subhani A, El-ghamry A, Changyong H, Jianming X. Effects of pesticides (herbicides) on soil microbial biomass: A review. Pakistan Journal of Biological Sciences 2000;3(5):705-9.
Supreeth M, Chandrashekar MA, Sachin N, Raju NS. Effect of chlorpyrifos on soil microbial diversity and its biotransformation by Streptomyces sp. HP-11.3. Biotech 2016;6(2):Article No.147.
Sviridova AV, Shushkova TV, Ermakova IT, Ivanova EV, Epiktetova DO, Leontievskya AA. Microbial degradation of glyphosate herbicides (review). Applied Biochemistry and Microbiology 2015;51(2):188-95.
Tatsinkou Fossi B, Tavea F, Ndjouenkeu R. Production and partial characterization of thermostable alpha amylase from ascomycete yeast strain isolated from starchy soil. African Journal of Biotechnology 2005;4(1):14-8.
Uqab B, Mudasi S, Nazir R. Review on bioremediation of pesticides. Journal of Bioremediation and Biodegradation 2016;7(3):Article No.1000343.
Williams ST, Goodfellow M, Alderson G, Wellington EMH, Sneath PHA, Sackin MJ. Numerical classification of Streptomyces and related genera. Journal of General Microbiology 1983;129(6):1743-813.
Xu ML, Gao Y, Li Y, Li X, Zhang H, Han XX, et al. Indirect glyphosate detection based on ninhydrine reaction and surface enhanced Raman scattering spectroscopy. Spectrochimica Acta - Part A: Molecular and Biomolecular Spectroscopy 2018;197:78-82.
Yu J, Jin B, Ji, Q, Wang H. Detoxification and metabolism of glyphosate by a Pseudomonas sp. via biogenic manganese oxidation. Journal of Hazardous Materials 2023;48(5):Article No. 130902.
Zhan H, Feng Y, Fan X, Chen S. Recent advances in glyphosate biodegradation. Applied Microbiology and Biotechnology 2018;102(12):5033-43.
Zhang H, Zhang Y, Hou Z, Wu X, Gao H, Sun F, et al. Biodegradation of triazine herbicide metribuzin by the strain Bacillus sp. N1. Journal of Environmental Science and Health, Part B 2014;49(2):79-86.
Zhang W, Li J, Zhang Y, Wu X, Zhou Z, Huang Y, et al. Characterization of a novel glyphosate-degrading bacterial species, Chryseobacterium sp. Y16C, and evaluation of its effects on microbial communities in glyphosate-contaminated soil. Journal of Hazardous Materials 2022;432:Article No. 128689.