Comparative of conventional dot blot hybridization and CARD dot blot hybridization for Salmonella detection in pork
Main Article Content
Abstract
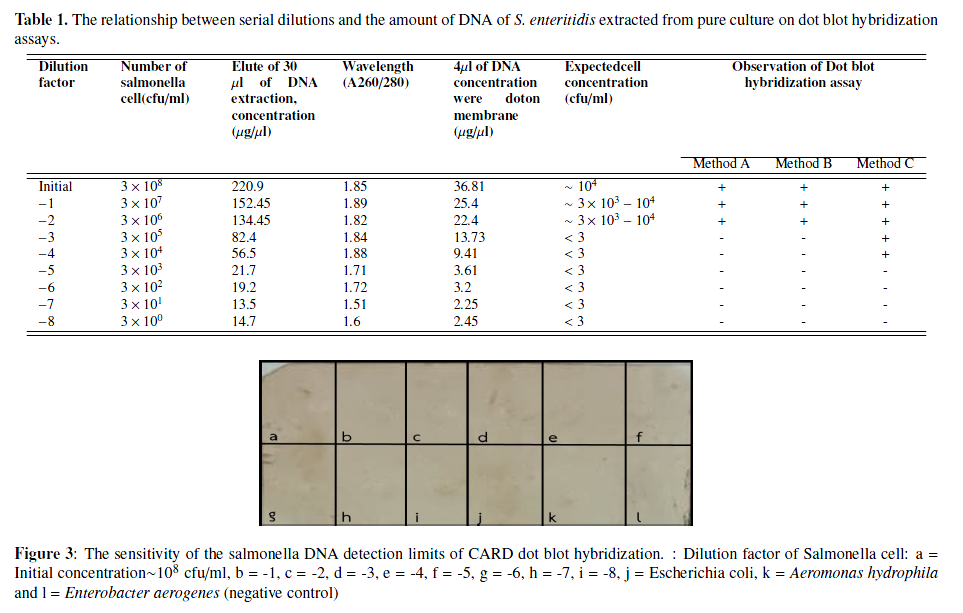
Dot blot hybridization assay was evaluated with Sal3 probe for rapid detection of Salmonella from pork samples. The Sal 3 probe (-5'OH) specificity with dot-blot hybridization found a DNA positive result of all salmonella serovars (S. typhimurium, S. enteritidis, S. vichow): while, there was negative result from 9 DNA samples of the negative control group. The conventional dot-blot hybridization methods (method A: System probe labeled DIG at 50-OH labeling DIG hybrids/ anti DIG-AP, detection with NBT/BCIP and method B: System probe labeled with biotin at 50-OH labeling biotin hybrids / streptavidin-HRP, detection with DAB) were compared with an application of catalyzed reporter deposition (CARD) to dot blot platform (method C: System probe labeled with biotin at 50-OH labeling biotin hybrids /1๐streptavidin-HRP /2๐streptavidin-HRP,+ system tyramide signal amplification (TSA), detection with DAB). The sensitivity of dot-blot hybridization methods for systems A, B and C were found C system has a high sensitivity for dot-blot hybridization method. The results were obtained at the lowest concentrations of 3 104 cfu / ml using a 2-day examination period. So, the separation processing of pathogens from meat samples is therefore essential. It is recommend that the use of appropriate DNA extraction kits or methods is critical for successful and valid CARD dot blot hybridization posed a challenge for salmonella detection on pork samples.
Article Details
References
CDC. Outbreaks Involving Salmonella(2018), https://www.cdc.gov/salmonella/outbreaks.html (accessed 27 April 2021).
EFSA (European Food Safety Authority), The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014, EFSA J.13 (12) 4329 (2015) 191.
R. S. Hendriksen, A. R. Vieira, S. Karlsmose, D. M. A. Lo Fo Wong, A. B. Jensen, H. C. Wegener, F. M. Aarestrup, Global monitoring of Salmonella serovar distribution from the World Health Organization Global foodborne infections network country data bank: Results of quality assured Laboratories from 2001 to 2007, Foodborne Pathogns and Disease 8(8) (2011) 1 – 14.
Z. Liang, B. Ke, X. Deng, J. Liang, L. Ran, L. Lu, D. He, Q. Huang, C. Ke, Z. Li, H. Yu, J. D. Klena, S. Wu, Serotypes, seasonal trends, and antibiotic resistance of nontyphoidal Salmonella from human patients in Guangdong Province, China, 2009 – 2012, BMC Infect. Dis. (2015) 55.
A. F`abrega, J. Vila, Salmonella enterica serovar typhimurium skills to succeed in the host: virulence and regulation, Clin. Microbiol. Rev. 26 (2013) 308 – 341.
EFSA, The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017, EFSa J 16 (2018).
EFSA, European Food Safety Authority. The European Union summary report on trends and sources of zoonoses, zoonotic agents and foodborne outbreaks in 2016, EFSA J. 15 (2017).
EFSA. European Food Safety Authority. Guidelines for reporting data on zoonoses, antimicrobial resistance and foodborne outbreaks using the EFSA data models for the data collection framework (DCF) to be used in 2017 for 2016 data, ISSN 2397-8325, (2016).
S. Angkititrakul, P. Tangkawattana, D. Sithigon , A. Polpakdee, D. Sithigon. Prevalence and antimicrobial resistance of salmonella isolated from been Khon Kaen municipality, Asia-Pacific Journal of Science and Technology 16 (2) (2011) 1 – 7.
C. Little, S. Walsh, L. Surman-Lee, K. Pathak,Y. Hall, E. De pinna, E. Threlfall, A. Maund, C. Chan, Salmonella contamination in non-UK produced shell eggs on retail scale in some regions of England, Weekly releases (1997 – 2007) 11 (47) (2006).
Y. Otomo, K. Abe, K. Odagiri, A. Shiroto, K. Takatori, Y. Hara-Kudo, Detection of Salmonella in spent hens and eggs associated with foodborne infections, Avian Diseases 51 (2007) 578 – 583.
M. Vieira-Pinto, M. Oliveira, J. Aranha, C. Martins, F. Bernardo, Influence of an enrichment step on Salmonella sp. detection by fluorescent in situ hybridization on pork samples, Food Control 19 (2008) 286 – 290.
B. Bisha, Fluorescence in situ hybridization-based detection of Salmonella spp. and Listeria monocytogenes in complex food matrices. Graduate Theses and Dissertations 10609 (2009).
D. Liu, Molecular detection of foodborne pathogen, CRC Press, 2009.
S. Ghazaleh, E. Zeinab, K. Hossein, Eciency of fluorescence in situ hybridization (FISH) method for the rapid detection of Salmonella in minced lamb meat: Method analysis and optimization, Journal of Microbiological Methods 175 (2020) 1 – 9.
O. Adebowale, L. Good, Development of a fixation-free fluorescence in situ hybridization for the detection of Salmonella species, Biology Methods and Protocols (2020) 1 – 12.
D. Arlai, S. Chuanchom, T. Sirinarumitr, Optimization of cell permeabilization for rapid detection of Salmonella in pork by FISH, Thai J Vet Med. 2015 45(1) (2012) 91 – 99.(in Thai)
M. Zerbini, M. Cricca, G. Gentilomi, S. Venturoli, G. Gallinella, M. Musiani, Tyramide signal amplification of biotinylated probe in dot-blot hybridization assay for the detection of parvovirus B19 DNA in serum samples, Clinica Chimica Acta 302(1-2) (2000) 79 – 87.
S. Nordentoft, H. Christensen, H. C. Wegener, Evaluation of a fluorescence-labelled oligonucleotide probe targeting 23S rRNA for in situ detection of Salmonella serovars in paraffinembedded tissue sections and their rapid identification in bacterial smears, J Clin Microbiol 35 (1997) 2642 – 2648.(in Thai)
M. M. Vieira-Pinto, M. Olivera, F. Bernardo, C. Martins, Evaluation of Fluorescent in situ hybridization (FISH) as a rapid screening method for detection of salmonella tonsils of slaughtered pig for consumption: a comparison with conventional culture method, Journal of Food Safety 25 (2005) 109 – 119.
B. O¨ rmerci, K. G. Linden, Development of a fluorescence in situ hybridization protocol for the identification of microorganisms associated with wastewater particles and flocs. J. Environ. Sci. and Health, Part A. 43 (2008) 1484 – 1488.
C. Almeida, N. F. Azevedo, R. M. Fernandes, C. W. Keevil, M. J. Vieira, Fluorescence in situ hybridization method using a peptide nucleic acid probe for identification of Salmonella spp. in a broad spectrum of samples, Appl Environ Microbiol 76 (2010) 4476 – 4485.
B. Bottari, G. E. Felis, E. Salvetti, A. Castioni, I. Campedelli, S.Torriani, V. Bernini, M. Gatti, E ective identification of Lactobacillus casei group species: genomebased selection of the gene mutL as the target of a novel multiplex PCR assay. Microbiology 163 (2017) 950 – 960.
G. H. Keller and M. M. Manak, DNA Probes. In: Stockton Press, New York (1989) 30 – 68.
K. Vaeteewootacharn, S. Sutra, S. Vaeteewootacharn, D. Sithigon, O. Jamjane, C. Chomvarin, C. Hahnvajanawong, N. Thongskulpanich, K. Thaewnon-giew, Salmonellosis and the food chain in Khon Kaen, Northeastern Thailand, Southeast Asian J Trop Med Public Health 36(1) (2005) 123 – 129.
S. Saleh, Salmonella Typhi, Paratyphi A, Enteritidis and Typhimurium core proteomes reveal di erentially expressed proteins linked to the cell surface and pathogenicity, PLoS Negl Trop Dis. 13(5) (2019).
L. Guillou, M-J. Chr´etiennot-Dinet, L. K. Medlin, H. Claustre, S. Loiseaux-de Goer, D. Vaulot, Bolidomonas,a new genus with two species belonging to new algal class, the Bolidophyceae Heterokonta, J. Phycol. 35 (1999) 368 – 381.
B. Karlsen, C. Cusack, E. Beensen, Microscopic and molecular methods for quantitative phytoplanktonanalysis. InIOC Manuals and Guides, No. 55 UNESCO: Paris, France, 2010.
Y. Kumar, R.Westram, S. Behrens, B. Fuchs, F. O. Gl¨ockner, R. Amann, H. Meier, W. Ludwig, Graphical representation of ribosomal RNA probe accessibility data using ARB software package.BMC Bioinform 6 (2005) 61.
M. Olivereira, F. Bernardo, Fluorescent In Situ Hybridization aplicado `a detec˜ao r´apida de Salmonella de origem alimentar eambiental, Rev. Port. Cien. Vet. 97 (2002) 81 – 85.
M. Vieira-Pinto, M. Oliveira, F. Bernardo, C. Martins, Rapid detection of Salmonella spp. in pork samples usingfluorescent in situ hybridization: a comparisonwith VIDAS (R)-SLM system and ISO 6579 cultural method. Arquivo Brasileiro deMedicina Veterinaria e Zootecnia 59 (2007) 1388 – 1393.
B. O¨ -rmerci, K. G. Linden, Development of a fluorescence in situhybridization protocol for the identification of microorganisms associated with wastewater particles and flocs, J. Environ. Sci. and Health Part A.43 (2008) 1484 – 1488.
N. D. Christensen, R. Kirnbauer, J. T. Schiller, S. -J. Ghim, R. Schlegel, A. B. Jenson, J. W. Kreider, Human papillomavirus types 6 and 11 have antigenically distinct strongly immunogenic conformationally dependent neutralizing epitopes, Virology 205 (1994) 329 – 335.
G. Van Camp, H. Fierens, P. Vandamme, H. Goossens, A. Huyghebaert, R. De Wachter, Identification of enteropathogenic Campylobacter species by oligonucleotide probes and polymerase chain reaction based on 16S rRNA genes, Syst Appl Microbiol 16 (1993) 30 – 36.
M. N. Bobrow, T. D. Harris, K. J. Shaughnessy, G. J. Litt, Catalyzed reporter deposition, a novel method of signal amplification: application to immunoassays, J. Immunol. Methods 125 (1989) 279 – 285.
H. Clay, L. Ramakrishnan, Multiplex fluorescent in situ hybridization in zebrafish embryos using tyramide signal amplification, Zebrafish 2 (2) (2005) 105 – 111.
A. U. Zaidi, H. Enomoto, J. Milbrandt, K. A. Roth, Dual fluorescent in situ hybridization and immunohistochemical detection with tyramide signal amplification, J Histochem Cytochem 48 (2000) 1369 – 1375.
A. Rohde, J. A. Hammerl, S. AI Dhouk, Detection of foodborne bacteria zoonoses by fluorescence in situ hybridization, Food Control 69 297 – 305.
I. G. Wilson, Inhibition and facilitation of nucleic acid amplification, Appl Envirn Microbiol (1997) 3741 – 3751.
M. C. Thomas, M. J. Shields, K. R. Hahn, T. W. Janzen, N. Goji, K. K. Amoako, Evaluation of DNA extraction methods for Bacillus anthracis spores isolated from spiked food samples, Journal of Applied Microbiology 115 (2013) 156 – 162.
N. Sajali, S. C. Wong, U. K. Hanapi, S. A. B Jamaluddin, N. A. Tasrip, N. A. Mohd Desa, The challenges of DNA extraction in different assorted food Matrices: A review, Journal of Food Science 83(10) (2018) 2409 – 2414.
I. Laube, J. Zagon, H. Broll, Quantitative determination of commercially relevant species in foods by real-time PCR, International Journal of Food Science and Technology 42 (2007) 336 – 341.
Z. Piskata, E. Pospisilova, G. Borilova, Comparative study of DNA extraction methods from fresh and processed yellowfin tuna muscle tissue. International Journal of Food, Properties 20 (1) (2017) 430 – 443.
Q. Zhang, S. Ishii, Improved simultaneous quantification of multiple waterborne pathogens and fecal indicator bacteria with the use of a sample process control, Water Research 137 (2018) 193 – 200.